Scientists use AI to unlock protein structures of hundreds of viruses for the first time
Published: 4 September 2024
Scientists are pioneering the use of machine-learning artificial intelligence software to investigate viruses, revealing never-before-seen viral mechanisms which yield immediate fundamental insights and pave the way for vaccine development
Scientists are pioneering the use of machine-learning artificial intelligence software to investigate viruses, revealing never-before-seen viral mechanisms which yield immediate fundamental insights and pave the way for vaccine development.
The research – published in Nature and led by the MRC-University of Glasgow Centre for Virus Research (CVR) in collaboration with the University of Sydney – use AI protein structure prediction to examine hundreds of species in the Flaviviridae, a large family of viruses that cause diseases such as Dengue, Zika and Hepatitis C.
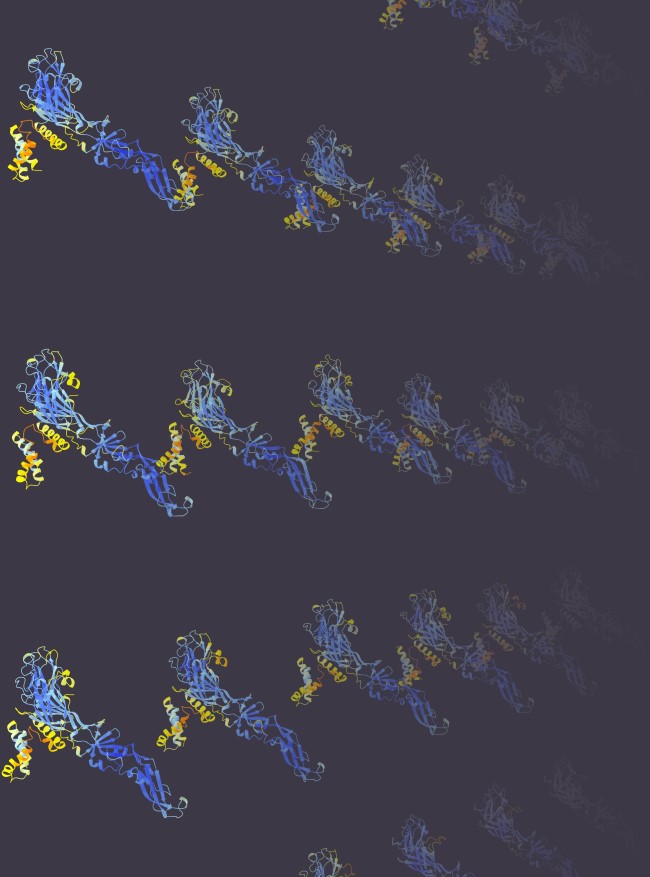
The work demonstrates a ‘super-charging’ of the scientific investigation into the evolution of viral proteins, uncovering the critically important entry mechanisms which explain how viruses get into the body and replicate in cells. This research not only provides various key biological insights, but also marks one of the first systematic application of protein structure prediction in virology, creating a resource for other investigators, and establishing a new paradigm for structure-informed exploration of virus evolution.
The AI technology, AlphaFold and ESMFold (developed by Google Deep Mind and Meta), was used to discover and classify the entry proteins of all the viruses tested – something which would be impossible to do with traditional methods.
The research authors believe the study to be an important step forward for future pandemic preparedness and current viral threats such as Mpox, for which scientists currently know very little about the entry proteins.
During the covid pandemic, scientists harnessed existing knowledge of the spike protein of SARS-CoV-2, the virus that causes COVID-19, to quickly develop vaccines. However, for many viruses, including Hepatitis C, the shape and mechanisms of the viral entry proteins are unknown.
This research demonstrates, for the first time, that Hepatitis C has a completely novel entry mechanism, unlike other viruses.
Dr Joe Grove, Senior Lecturer at the MRC-University of Glasgow Centre for Virus Research, said: “We are hugely excited by the results this study offers us. We are one of the first research groups to apply this AI technology at scale to viruses, and the results have huge implications for understanding how viruses get into our bodies and replicate, something which is critically important for future vaccine development, pandemic preparedness and furthering our knowledge of potential spillover viruses.
“By discovering more about the entry proteins on the outside of viruses, as we’ve done, we can better understand the fundamentals of viral biology, which in turn can guide development of drugs or vaccines.
“We are particularly excited about the discoveries about the entry proteins of Hepatitis C. There is currently no vaccine for hepatitis C, so we are hopeful that our new understanding of this virus’s entry mechanism will help lead to the development of a new vaccine.
“Going forward we want to use this technology to scale up our research to thousands of viruses. By doing this we can build foundational knowledge to inform our responses to existing and new viral diseases”
The study, ‘Mapping glycoprotein structure reveals Flaviviridae evolutionary history,’ is published in Nature. The work was funded by the Wellcome Trust, Royal Society and MRC.
Enquiries: ali.howard@glasgow.ac.uk or elizabeth.mcmeekin@glasgow.ac.uk
First published: 4 September 2024

